genomad|how to cite gnomad : Manila Insights - GitHub - apcamargo/genomad: geNomad: Identification of mobile .
Learn about the different types of earths and grounds, and how they apply to everyday electronic circuits.
PH0 · what is gnomad sg org
PH1 · how to cite gnomad
PH2 · gnomad v2.1.1
PH3 · gnomad genome browser
PH4 · gnomad download
PH5 · gnomad browser
PH6 · genome browser
PH7 · build your own van home
PH8 · Iba pa
3. Players must check for the accuracy of printed data on their ticket, including bet type, bet amount, the draw date and the numbers they want to play. Swertres Result Today, July 12, 2024 Updates Monitor 3D Lotto, Swertres Result Today
genomad*******geNomad is a tool to identify viruses and plasmids in sequencing data. Learn how to install, use, and cite geNomad, and explore its web app and documentation.Issues 2 - GitHub - apcamargo/genomad: geNomad: Identification of mobile .Pull requests - GitHub - apcamargo/genomad: geNomad: .
Actions - GitHub - apcamargo/genomad: geNomad: Identification of mobile .
GitHub is where people build software. More than 100 million people use .Insights - GitHub - apcamargo/genomad: geNomad: Identification of mobile .
Changed. The mmseqs search command has been replaced by a two-step .
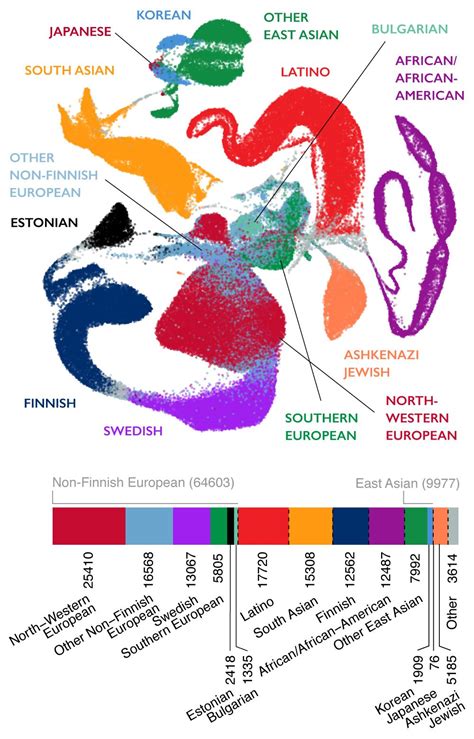
The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both .
Abstract. Identifying and characterizing mobile genetic elements in sequencing data is essential for understanding their diversity, ecology, biotechnological .
gnomAD is a database that collects and harmonizes exome and genome sequencing data from various projects and makes summary data available. It has two versions: v2.1.1 .Here we introduce geNomad, a classification and annotation framework that combines information from gene content and a deep neural network to identify sequences of .
geNomad is a fast and accurate tool that can find and classify mobile genetic elements from nucleotide sequences. It also provides functional annotation and taxonomic assignment for the identified viruses and . gnomAD v4.0 includes data from 807,162 individuals, with exome and genome sequencing data from 730,947 and 76,215 individuals, respectively. Learn .genomad how to cite gnomad The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and .gnomAD aggregates and analyses 15,708 whole genomes and 125,748 exomes from diverse populations. It provides a catalogue of genetic variants, their functional impact, . Here, we introduce geNomad, a classification and annotation framework that combines information from gene content and a deep neural network to identify .
gnomAD. The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider scientific community.The command to execute geNomad is structured like this: So, to run the full geNomad pipeline ( end-to-end command), taking a nucleotide FASTA file ( GCF_009025895.1.fna.gz) and the database ( genomad_db) as input, we will execute the following command: The results will be written inside the genomad_output directory.We would like to show you a description here but the site won’t allow us. gnomAD v2.1 comprises a total of 16mln SNVs and 1.2mln indels from 125,748 exomes, and 229mln SNVs and 33mln indels from 15,708 genomes. In addition to the 7 populations already present in gnomAD 2.0.2, this release now breaks down the non-Finnish Europeans and East Asian populations further into sub-populations. The . The gnomAD dataset of over 270 million variants is publicly available (https://gnomad.broadinstitute.org), and has already been widely used as a resource for estimates of allele frequency in the . gnomAD v4.1. We have released gnomAD v4.1, an update to our latest major release. This update fixes the allele number issue in gnomAD v4.0 previously described here and critically adds two new functionalities: joint allele number across all called sites in the exomes and genomes and a new flag indicating when the frequencies . 本文内容. 数据源. 数据量和更新频率. 存储位置. 数据访问. 显示另外 3 个. 基因组聚合数据库 (gnomAD) 是一种由国际研究人员联盟开发的资源,其目标是聚合和协调各种大型测序项目的外显子组和基因组测序数据,并为更广泛的科学社区提供汇总数据。.
The gnomAD v2 call set contains fewer whole genomes than v3.1, but also contains a very large number of exomes that substantially increase its power as a reference in coding regions. Therefore gnomAD v2 is still our recommended dataset for most coding regions analyses. However, gnomAD v3.1 represents a very large increase in the number of .
We would like to show you a description here but the site won’t allow us.
Next post: gnomAD v2.1. The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider . The gnomAD resource illustrates both the power and the challenges of interpreting human biology using large‐scale genomic datasets. The sheer size of gnomAD makes it possible to obtain accurate estimates of allele frequency extending down to incredibly rare variation, and to explore the patterns of variation across genes and .genomad The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider scientific community. geNomad is an annotation and classification framework that combines and builds on two standard techniques for identifying viruses and plasmids. Until now, most other tools have focused on identifying only specific plasmids or viruses. geNomad combines a broad scope, targeting all known groups of viruses and plasmids. .Abstract. Reference population databases are an essential tool in variant and gene interpretation. Their use guides the identification of pathogenic variants amidst the sea of benign variation present in every human genome, and supports the discovery of new disease-gene relationships. The Genome Aggregation Database (gnomAD) is currently . The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider scientific community.how to cite gnomadThe geNomad pipeline #. The geNomad pipeline. #. When you execute the genomad end-to-end command, geNomad runs a series of modules sequentially to produce the final output, which contains the identified plasmids and viruses in the input FASTA file. It is possible to execute the modules sequentially, which allows you to use some advanced .
Identifying and characterizing mobile genetic elements (MGEs) in sequencing data is essential for understanding their diversity, ecology, biotechnological applications, and impact on public health. Here, we introduce geNomad, a classification and annotation framework that combines information from gene content and a deep neural network to .

Leveraging geNomad’s speed and scalability, we processed over 2.7 trillion base pairs of sequencing data, leading to the discovery of millions of viruses and plasmids that are available through .
Unlimited Free Algorithmic Mastering by BandLab. Fast, High Quality Online Mastering. Instantly master your tracks with the world’s leading online mastering service. Hear the difference mastering can make with the fastest, best sounding, and free artist-driven Mastering tool.For the most comprehensive collection of FREE porn categories online, visit YouPorn! Browse through our selection of free sex videos from popular XXX categories, such as Lesbian, Mature, Anal, 18+ Teen, Amateur, MILF and Threesome.
genomad|how to cite gnomad